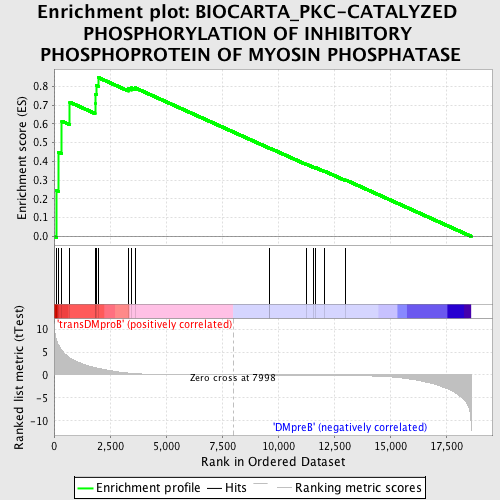
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | BIOCARTA_PKC-CATALYZED PHOSPHORYLATION OF INHIBITORY PHOSPHOPROTEIN OF MYOSIN PHOSPHATASE |
| Enrichment Score (ES) | 0.84740967 |
| Normalized Enrichment Score (NES) | 1.6034181 |
| Nominal p-value | 0.003992016 |
| FDR q-value | 0.17972699 |
| FWER p-Value | 0.756 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARHGEF5 | 12025 | 106 | 7.710 | 0.2453 | Yes | ||
| 2 | MYLK | 22778 4213 | 202 | 6.449 | 0.4501 | Yes | ||
| 3 | VAV1 | 23173 | 347 | 5.300 | 0.6149 | Yes | ||
| 4 | PKN1 | 11567 6799 | 704 | 3.692 | 0.7159 | Yes | ||
| 5 | GNA13 | 20617 | 1827 | 1.605 | 0.7079 | Yes | ||
| 6 | CAMK2B | 20536 | 1834 | 1.586 | 0.7592 | Yes | ||
| 7 | ROCK1 | 5386 | 1898 | 1.517 | 0.8052 | Yes | ||
| 8 | GNB1 | 15967 | 1980 | 1.431 | 0.8474 | Yes | ||
| 9 | ARHGEF1 | 1952 18343 3996 | 3312 | 0.395 | 0.7887 | No | ||
| 10 | MYL2 | 16713 | 3453 | 0.339 | 0.7922 | No | ||
| 11 | GNA12 | 9023 | 3615 | 0.280 | 0.7927 | No | ||
| 12 | GNAQ | 4786 23909 3685 | 9625 | -0.023 | 0.4704 | No | ||
| 13 | GNGT1 | 17533 | 9636 | -0.023 | 0.4706 | No | ||
| 14 | CACNA1A | 4461 | 11245 | -0.051 | 0.3858 | No | ||
| 15 | PPP1R14A | 18310 3822 | 11597 | -0.060 | 0.3689 | No | ||
| 16 | PRKCA | 20174 | 11681 | -0.062 | 0.3664 | No | ||
| 17 | ARHGAP5 | 4412 8625 | 12065 | -0.074 | 0.3483 | No | ||
| 18 | PLCB1 | 14832 2821 | 13019 | -0.120 | 0.3009 | No |

